Generating 2D Gating Hierarhcy from Clustered Cytometry Data
Our algorithm cluster-to-gates(C2G) can visualize a clustered cytometry data in 2D gating hierarchy. The overall input to C2G are two variables: "data" and "label". "data" is a N-by-M matrix where N is the number of cells and M is the number of markers. "label" is a N-by-1 matrix and this matrix represent which cluster each cell belongs to (0 for ungated cells). In this page, we will demonstrate how to use C2G to generate gating hierarchy on a simulated data and on a CyTOF dataset that are manually gated. The software and testing data can be downloaded here .
Contents
- Initialization and load the simulated data
- Visualize the simulated data and pre-clustered simulated data in 3D
- Run the analysis on the simulated data
- Load a moderate-sized CyTOF dataset with ~10,000 cells
- Generate gating hierarchy for manually gated populations
- Generate gating hierarchy for K-means defined populations (K=10)
- Apply C2G to large CyTOF dataset with ~140,000 cells
- Reference
Initialization and load the simulated data
This simulated data set contains 20000 cells, which consists of 5 populations.Two of the populations are considered as target and labeled as 1 and 2. The cells in other populations are labeled 0 which means "unlabeled". A visualization of the original data is also shown in the following sections.
addpath('src') addpath('libs') load('testdata/simulated.mat','data','label'); markernames ={'Marker 1','Marker 2','Marker 3'}; fprintf('Size of "data" is %d-by-%d\n',size(data)); fprintf('Size of "label" is %d-by-%d\n',size(label));
Size of "data" is 20000-by-3 Size of "label" is 20000-by-1
Visualize the simulated data and pre-clustered simulated data in 3D
In real application, this section is not necessay.
col = [0.8 0.8 0.8;hsv(length(unique(label))-1)]; figure('Position',[680 478 560 420]); scatter3(data(:,1),data(:,2),data(:,3),1,col(label+1,:)); xlabel('Marker 1') ylabel('Marker 2') zlabel('Marker 3') axis([-5 15 -5 15 -5 15]);
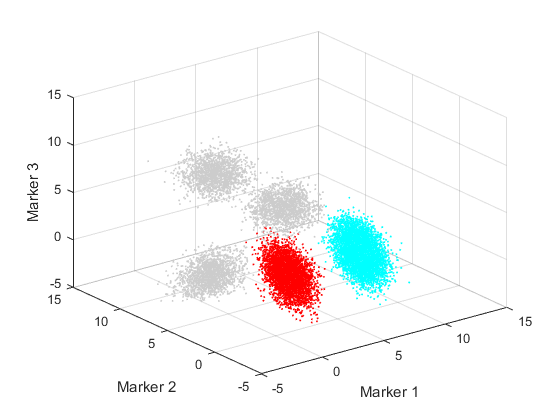
Run the analysis on the simulated data
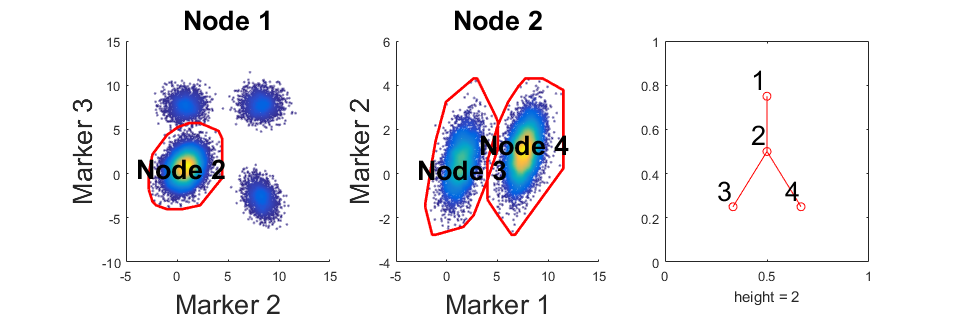
This section performs the analysis on the simulated data. If your data has no "unlabeled" cells, precluster step can be skipped. If you skip precluster, the second and third parameter in function "C2G" should be the same.
% Precluster the simulated data rng(9464); preclustered_label = cluster_ungated(data,label); % Call main part of the program and return a object m that store the % results. m = C2G(data,preclustered_label,label,'markernames',markernames); % Draw the obtained gating hierarchy m.view_gates(data,markernames,'n_lines',1,'onepanel',true); % Show statistics outtable = m.show_f_score(label);
Step: 0 [100%]
[Selected] marker pair Marker 2 and Marker 3
Population Gate TP FP FN F-score
1 1 5411 14589 0 0.426
2 1 9166 10834 0 0.629
Normalized Mutual Information = 0.321
Step: 1 [100%]
[Selected] marker pair Marker 1 and Marker 2
Population Gate TP FP FN F-score
1 2 5411 9183 0 0.541
2 2 9166 5428 0 0.772
Normalized Mutual Information = 0.736
Step: 2 [100%]No further separation. Node 3 is gate for cell population 1
Step: 3 [100%]No further separation. Node 4 is gate for cell population 2
Population Gate TP FP FN F-score
1 3 5411 18 0 0.998
2 4 9166 1 0 1.000
Normalized Mutual Information = 0.996
Load a moderate-sized CyTOF dataset with ~10,000 cells
This is a dataset about T cell signaling (Krishnaswamy et al, Science, 2014). It can be downloaded from here. This section will load multiple fcs files. Each fcs file correspond to one cell population manually gated by the authors. C2G will automatically generate cell labels based on fsc files. FCS files are store in the "testdata" folder. "CD4_Effmen.fcs", "CD4_naive.fcs", and "CD8_naive.fcs" are cells of target populations (manually defined) and "ctr.fcs" contain all cells. In this example, we only use surface protein markers.
clear close all addpath('src') addpath('libs') fdname = 'testdata'; [ori_data,ori_l,ori_markers]=load_mul_fcs(fdname,'ctr.fcs'); surface_idx = [3 4 6 8 9 11 12 13 22 24 25 27]; data = ori_data(:,surface_idx); markers = ori_markers(surface_idx); n_markers = length(markers);
Generate gating hierarchy for manually gated populations
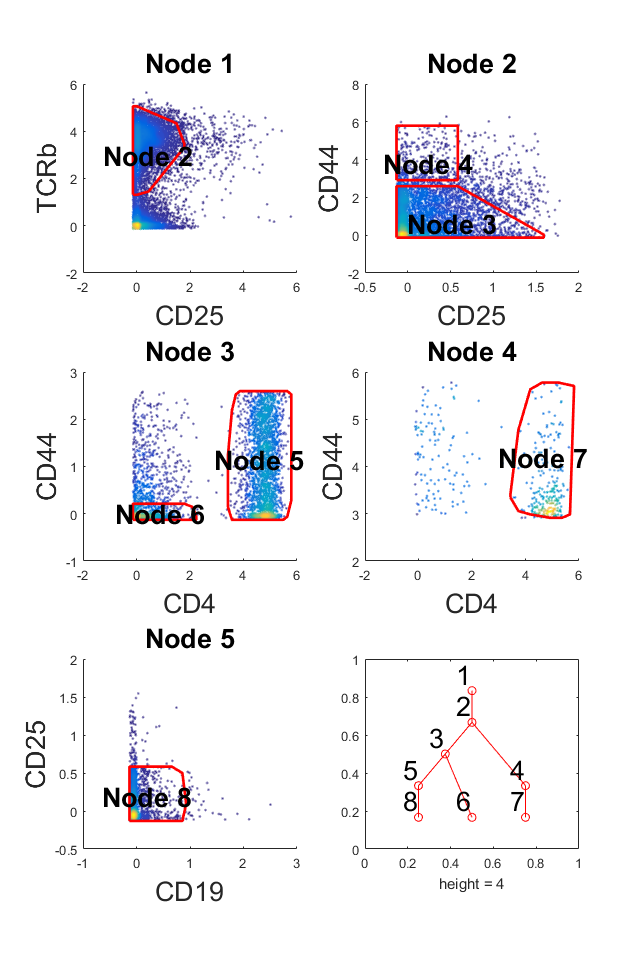
In this section, we'll use the data loaded from previous section and treat the three manually defined cell populations as target populations. Here, number of "unlabeled" cells is around two-fold of cells in target populations. The results (the table below) show that C2G can capture manually defined populations at very high accuracy.
% Precluster the ungated cells rng(9464) label = cluster_ungated(data,ori_l); % Perform the anlysis m_ori = C2G(data,label,ori_l,'markernames',markers); % Visualize the results m_ori.view_gates(data,markers,'n_lines',3,'ignore_small',0,'onepanel',true); m_ori.show_f_score(ori_l);
Population Gate TP FP FN F-score
1 7 216 13 8 0.954
2 8 2267 30 78 0.977
3 6 488 46 37 0.922
Normalized Mutual Information = 0.893
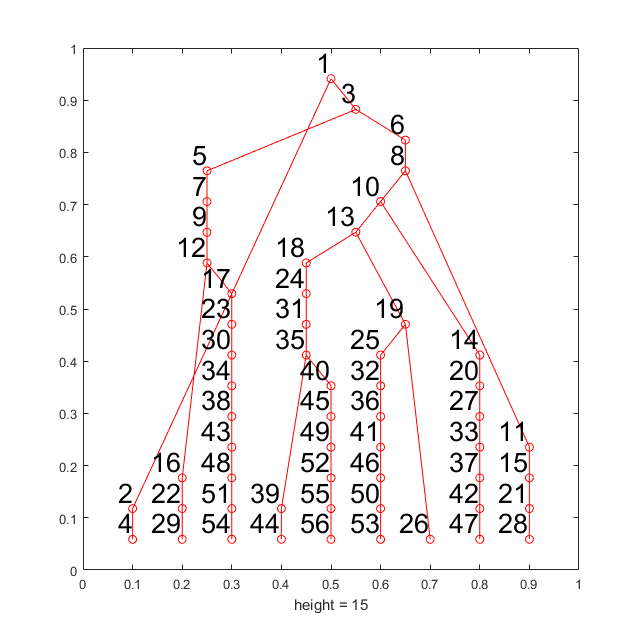
Generate gating hierarchy for K-means defined populations (K=10)
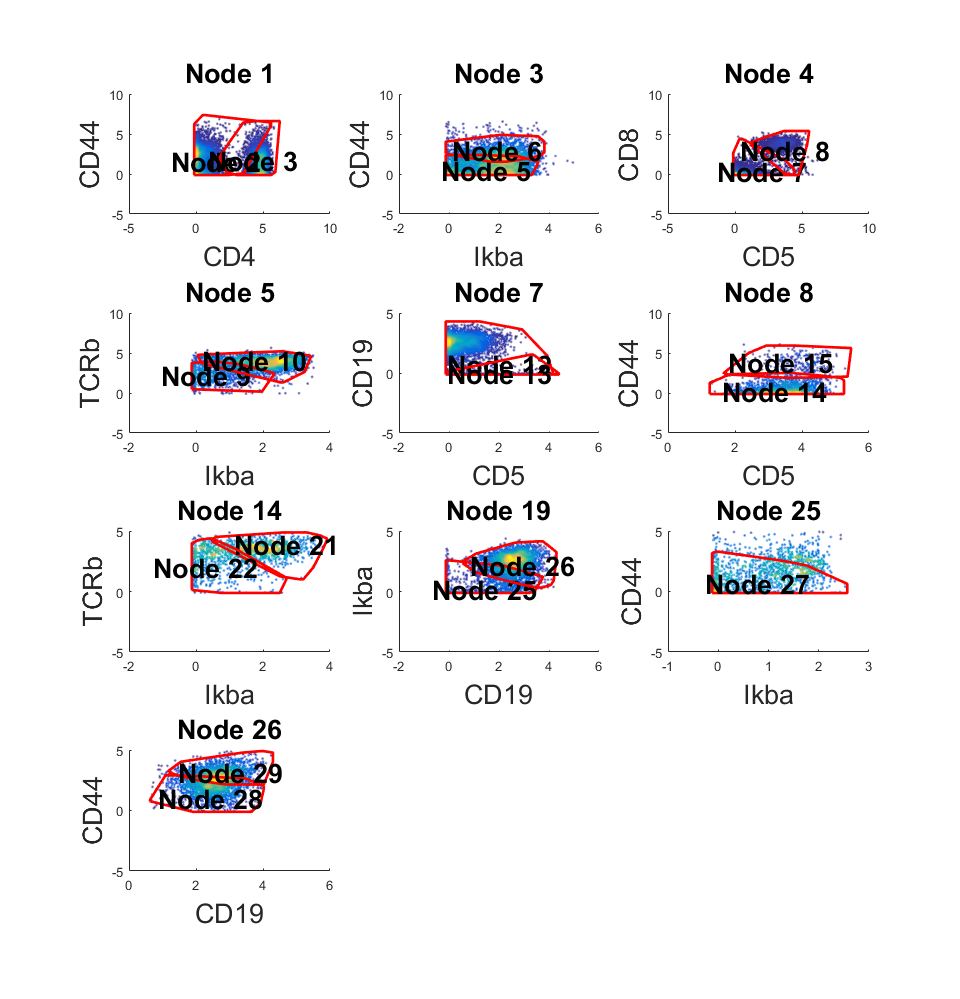
In this section, we will use the same CyTOF data used in previous section. The cell labels are defined by k-means algorithm where k equal to 10. All of the 10 clusters will be considered as target populations, in other words,there're no "unlabeled" cells.
rng(9464) km_l = kmeans(data,10); new_km_l = km_l; % Since all populiations is known, no need to pre-cluster % Perform the anlysis m_km = C2G(data,km_l,km_l,'markernames',markers); % Visualize the results m_km.view_gates(data,markers,'n_lines',4,'ignore_small',300); m_km.show_f_score(km_l);
Population Gate TP FP FN F-score
1 27 746 155 42 0.883
2 16 798 139 115 0.863
3 32 1204 176 85 0.902
4 24 1853 243 277 0.877
5 31 1418 161 74 0.923
6 18 1163 165 208 0.862
7 21 714 27 38 0.956
8 20 138 12 18 0.902
9 22 609 38 46 0.935
10 23 192 15 26 0.904
Normalized Mutual Information = 0.780
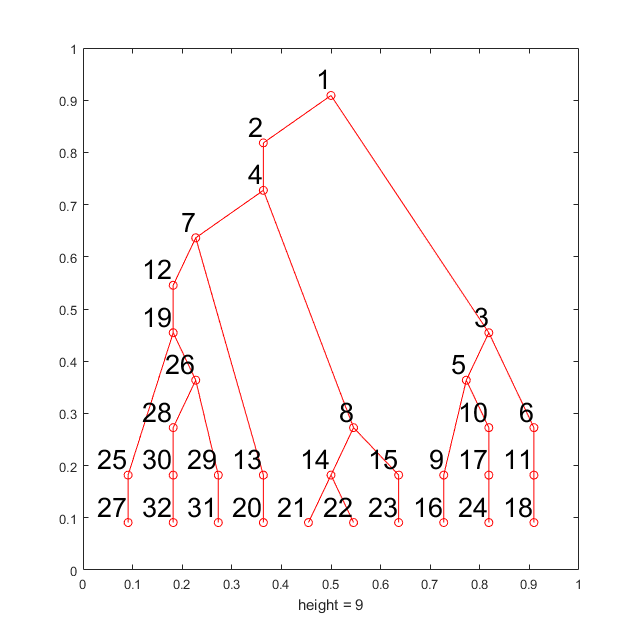
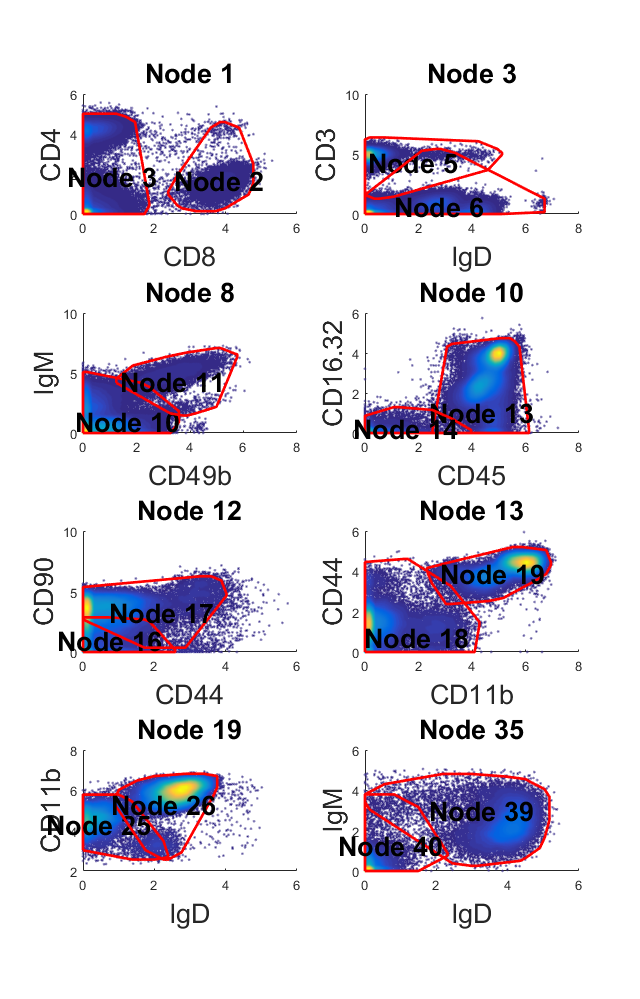
Apply C2G to large CyTOF dataset with ~140,000 cells
This is a larger CyTOF dataset with 140k cells and 21 protein markers (Spitzer, Matthew H., et al, Science, 2015 ). This data can be download for free from here. (The dataset is hosted on Cytobank and you need to register to download it) This dataset is clustered by k-means where k equal to 10. All of the 10 k-means defined clusters are treated as target populations. This part will take around 10 minutes on a desktop.
[ori_data, marker] = readfcs_v2('testdata/bigdata/TIN_BLD1_Untreated_Day3.fcs'); % Transform the CyTOF data = flow_arcsinh(ori_data,5); % Select protein markers marker_idx = [10,16,17,18,19,21,22,24,29,31,32,33,39,40,41,46,47,49,50,51,52]; d = data(marker_idx,:)'; rng(9464); label = kmeans(d,10); % Perform the anlysis tic;m = C2G(d, label, label,'markernames',marker(marker_idx));toc; % Visualize the results w = warning ('off','all'); m.view_gates(d,marker(marker_idx),'ignore_small',3000,'n_lines',4); warning(w); m.show_f_score(label);
Population Gate TP FP FN F-score
1 29 15390 534 1172 0.947
2 47 6683 255 544 0.944
3 26 10863 14070 1338 0.585
4 44 19313 85 822 0.977
5 53 9833 301 1164 0.931
6 26 13421 11512 784 0.686
7 4 13324 35 442 0.982
8 54 29331 833 1526 0.961
9 28 6993 460 398 0.942
10 56 7804 218 726 0.943
Normalized Mutual Information = 0.859
Reference
Krishnaswamy, Smita, Matthew H. Spitzer, Michael Mingueneau, Sean C. Bendall, Oren Litvin, Erica Stone, Dana Pe’er, and Garry P. Nolan. "Conditional density-based analysis of T cell signaling in single-cell data." Science 346, no. 6213 (2014): 1250689.
Spitzer, Matthew H., Pier Federico Gherardini, Gabriela K. Fragiadakis, Nupur Bhattacharya, Robert T. Yuan, Andrew N. Hotson, Rachel Finck et al. "An interactive reference framework for modeling a dynamic immune system." Science 349, no. 6244 (2015): 1259425.